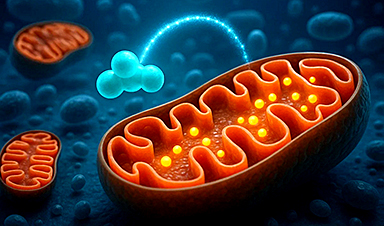
Researchers from the LMS and LMB have discovered how the D2-I protein complex identifies and repairs DNA damage, a breakthrough that promises to enhance cancer treatments by improving our understanding of DNA repair pathways. This collaboration could pave the way for more effective therapies by targeting the mechanisms that cancer cells use to resist treatment.
A collaboration between researchers at the UK’s two core-funded Medical Research Council Institutes—the Laboratory of Medical Sciences (LMS) in London and the Laboratory of Molecular Biology (LMB) in Cambridge—has unraveled a decades-old mystery, potentially leading to improved cancer treatments in the future.
The work, which uncovered the basic mechanism of how one of our most vital DNA repair systems recognizes DNA damages and initiates their repair, has eluded researchers for many years. Using cutting-edge imaging techniques to visualize how these DNA repair proteins move on a single molecule of DNA, and electron microscopy to capture how they “lock-on” to specific DNA structures, this research opens the way to more effective cancer treatments.
The collaboration between the laboratories of Professor David Rueda (LMS) and Dr Lori Passmore (LMB) has been a brilliant example of how #teamscience can bear fruitful results and underscores the importance of these two institutes in driving forward research that unlocks the fundamental mechanisms of biology which will underpin the future translation of that work into improvements in human health.
A single molecule of DNA (not directly visible) is captured using microscopic beads (the large circles). Each of the red, green, or yellow dots moving between the beads represents a FANCD2I-FANCI protein complex sliding along the DNA molecule, monitoring it for damage. Credit: MRC Laboratory of Medical Sciences
Unraveling the DNA Repair Mechanism
The researchers were working on a DNA repair pathway, known as the Fanconi Anaemia [FA] pathway, which was identified more than twenty years ago. DNA is constantly damaged throughout our lives by environmental factors including UV light from the sun, alcohol use, smoking, pollution, and exposure to chemicals. One way in which DNA becomes damaged is when it is “cross-linked”, which stops it being able to replicate and express genes normally. In order to replicate itself and to read and express genes, the two strands of the DNA double helix first has to unzip into single strands. When DNA is cross-linked, the “nucleotides” (the “steps” in the double-helix ladder of DNA) of the two strands become stuck together, preventing this unzipping.
The accumulation of DNA damages including cross-linking can lead to cancer. The FA pathway is active throughout our lives and identifies these damages and repairs them on an ongoing basis. Individuals who have mutations that make this pathway less effective are far more susceptible to cancers. Although the proteins involved in the FA pathway were discovered some time ago, a mystery remained over how they identified the cross-linked DNA and started the process of DNA repair.
The team from the MRC LMS sister institution, the LMB in Cambridge, led by Lori Passmore, had previously identified that the FANCD2-FANCI (D2-I) protein complex, which acts in one of the first steps of the FA pathway, clamps onto DNA, thereby initiating DNA repair at crosslinks. However, key questions remained: how does D2-I recognize crosslinked DNA, and why is the D2-I complex also implicated in other types of DNA damage?
The research, published in the journal Nature, used a combination of cutting-edge scientific techniques to show that the D2-I complex slides along the double-stranded DNA, monitoring its integrity, and has also elegantly visualized how it recognizes where to stop, allowing the proteins to move and lock together at that point to initiate DNA repair.
Advanced Techniques Shed Light on Molecular Interactions
Artur Kaczmarczyk and Korak Ray in David Rueda’s Single Molecule Imaging group, working with Pablo Alcón in Lori Passmore’s group, used a state-of-the-art microscopy technique known as “correlated optical tweezers and fluorescence imaging” to explore how the D2-I complex slides along a double-stranded DNA molecule.
Using optical tweezers, they could catch a single DNA molecule between two beads, which allowed them to precisely manipulate the DNA and incubate it with chosen proteins. Using fluorescently labeled D2-I and single-molecule imaging, they observed how individual D2-I complexes bind to and slide along DNA, scanning the double helix. They discovered that rather than recognizing the crosslink between the two strands of DNA directly, the FA clamp instead stops sliding when it reaches a single-stranded DNA gap, a region where one of the two strands of DNA is missing.
The video shows the FANCD2-FANCI complex clamping to DNA in order to repair it. Credit: MRC Laboratory of Medical Sciences, MRC Laboratory for Molecular Biology
Using cryo-electron microscopy, a powerful technique which can visualize proteins at a molecular level, the researchers next determined the structures of the D2-I complex both in its sliding position and stalled at the junction between single-stranded and double-stranded DNA. This revealed that the contacts D2-I makes with this single-stranded–double-stranded DNA junction are distinct from the contacts it makes with double-stranded DNA alone. This allowed them to identify a specific portion of the FANCD2 protein, called the “KR helix” that they showed in their single-molecule imaging experiments is critical for recognizing and stalling at the single-stranded DNA gaps.
Working with Guillaume Guilbaud and Julian Sale in the LMB’s PNAC Division, and Themos Liolios and Puck Knipscheer at the Hubrecht Institute, Netherlands, they further showed that the D2-I complex’s ability to stall at these junctions using the KR helix is critical for DNA repair by the FA pathway.
When DNA normally replicates in our cells, it unzips the two DNA strands and copies each single strand. This creates a ‘replication fork’ where the original DNA strands are unwound and new double-stranded DNA is formed on each strand. However, when this fork reaches a DNA crosslink, the strands cannot be unzipped, stalling the usual DNA replication process. This stalled replication fork thus contains exposed single-stranded gaps where the DNA has been unwound but not replicated. This research has shown that it is these junctions between single- and double-stranded DNA at the stalled replication fork that the D2-I protein complex latches tightly onto.
Implications for Cancer Treatment and Beyond
Not only does this allow D2-I complex to bring other FA pathway proteins to the DNA crosslink to initiate repair, but it also anchors the remaining double-stranded DNA, protecting the stalled “replication fork” from enzymes in the cell that would chew up the exposed end of the DNA strand and further damage the DNA. This work has shown that it is DNA structures within the replication fork that stalls as a result of cross-linked DNA, rather than the cross-linked DNA itself, that triggers the D2-I complex to stop sliding and clamp on to DNA to initiate repair. These stalled replication forks appear in many types of DNA damage, explaining the broad role of the D2-I complex in other forms of DNA repair as well as via the FA pathway.
Understanding the process of DNA repair, and, importantly, why it fails, holds huge importance as DNA damage is a key factor in many diseases. Critically, many cancer drugs, for example, Cisplatin, work by inducing such serious cellular damage to cancer cells that they stop dividing and die. In such cases, DNA repair pathways—such a vital physiological process in normal life—can be hijacked by cancer cells that use them to resist the effects of chemotherapy drugs. Understanding the mechanistic basis of the first step in the DNA repair pathway may lead to ways of sensitizing patients so that cancer drugs can be more effective in the future.
Reference: “FANCD2–FANCI surveys DNA and recognizes double- to single-stranded junctions” by Pablo Alcón, Artur P. Kaczmarczyk, Korak Kumar Ray, Themistoklis Liolios, Guillaume Guilbaud, Tamara Sijacki, Yichao Shen, Stephen H. McLaughlin, Julian E. Sale, Puck Knipscheer, David S. Rueda and Lori A. Passmore, 31 July 2024, Nature.
DOI: 10.1038/s41586-024-07770-w
This work was funded by UKRI MRC, the Wellcome Trust, the European Research Council, and the EMBO.
News
Global Nanomaterial Regulation: A Country-by-Country Comparison
Nanomaterials are materials with at least one dimension smaller than 100 nanometres (about 100,000 times thinner than a human hair). Because of their tiny size, they have unique properties that can be useful in [...]
Pandemic Potential: Scientists Discover 3 Hotspots of Deadly Emerging Disease in the US
Virginia Tech researchers discovered six new rodent carriers of hantavirus and identified U.S. hotspots, highlighting the virus’s adaptability and the impact of climate and ecology on its spread. Hantavirus recently drew public attention following reports [...]
Studies detail high rates of long COVID among healthcare, dental workers
Researchers have estimated approximately 8% of Americas have ever experienced long COVID, or lasting symptoms, following an acute COVID-19 infection. Now two recent international studies suggest that the percentage is much higher among healthcare workers [...]
Melting Arctic Ice May Unleash Ancient Deadly Diseases, Scientists Warn
Melting Arctic ice increases human and animal interactions, raising the risk of infectious disease spread. Researchers urge early intervention and surveillance. Climate change is opening new pathways for the spread of infectious diseases such [...]
Scientists May Have Found a Secret Weapon To Stop Pancreatic Cancer Before It Starts
Researchers at Cold Spring Harbor Laboratory have found that blocking the FGFR2 and EGFR genes can stop early-stage pancreatic cancer from progressing, offering a promising path toward prevention. Pancreatic cancer is expected to become [...]
Breakthrough Drug Restores Vision: Researchers Successfully Reverse Retinal Damage
Blocking the PROX1 protein allowed KAIST researchers to regenerate damaged retinas and restore vision in mice. Vision is one of the most important human senses, yet more than 300 million people around the world are at [...]
Differentiating cancerous and healthy cells through motion analysis
Researchers from Tokyo Metropolitan University have found that the motion of unlabeled cells can be used to tell whether they are cancerous or healthy. They observed malignant fibrosarcoma [...]
This Tiny Cellular Gate Could Be the Key to Curing Cancer – And Regrowing Hair
After more than five decades of mystery, scientists have finally unveiled the detailed structure and function of a long-theorized molecular machine in our mitochondria — the mitochondrial pyruvate carrier. This microscopic gatekeeper controls how [...]
Unlocking Vision’s Secrets: Researchers Reveal 3D Structure of Key Eye Protein
Researchers have uncovered the 3D structure of RBP3, a key protein in vision, revealing how it transports retinoids and fatty acids and how its dysfunction may lead to retinal diseases. Proteins play a critical [...]
5 Key Facts About Nanoplastics and How They Affect the Human Body
Nanoplastics are typically defined as plastic particles smaller than 1000 nanometers. These particles are increasingly being detected in human tissues: they can bypass biological barriers, accumulate in organs, and may influence health in ways [...]
Measles Is Back: Doctors Warn of Dangerous Surge Across the U.S.
Parents are encouraged to contact their pediatrician if their child has been exposed to measles or is showing symptoms. Pediatric infectious disease experts are emphasizing the critical importance of measles vaccination, as the highly [...]
AI at the Speed of Light: How Silicon Photonics Are Reinventing Hardware
A cutting-edge AI acceleration platform powered by light rather than electricity could revolutionize how AI is trained and deployed. Using photonic integrated circuits made from advanced III-V semiconductors, researchers have developed a system that vastly [...]
A Grain of Brain, 523 Million Synapses, Most Complicated Neuroscience Experiment Ever Attempted
A team of over 150 scientists has achieved what once seemed impossible: a complete wiring and activity map of a tiny section of a mammalian brain. This feat, part of the MICrONS Project, rivals [...]
The Secret “Radar” Bacteria Use To Outsmart Their Enemies
A chemical radar allows bacteria to sense and eliminate predators. Investigating how microorganisms communicate deepens our understanding of the complex ecological interactions that shape our environment is an area of key focus for the [...]
Psychologists explore ethical issues associated with human-AI relationships
It's becoming increasingly commonplace for people to develop intimate, long-term relationships with artificial intelligence (AI) technologies. At their extreme, people have "married" their AI companions in non-legally binding ceremonies, and at least two people [...]
When You Lose Weight, Where Does It Actually Go?
Most health professionals lack a clear understanding of how body fat is lost, often subscribing to misconceptions like fat converting to energy or muscle. The truth is, fat is actually broken down into carbon [...]